Proteomics Core Facility

The Proteomics Core Facility (PCF) provides top-notch mass spectrometry-based proteomic technologies and relevant training to the EPFL Life Sciences community and collaborators.
Proteins are essential building blocks of all living organisms. The collection of proteins expressed by a cell, tissue, or organism at a given time is collectively referred as the proteome. Proteomics represents the effort to establish the identities, quantities, structures, and functions of all proteins in a given model and how these properties vary in different conditions. Mass-spectrometry-based proteomics is a highly successfully technique for the identification and characterization of proteins and proteomes.
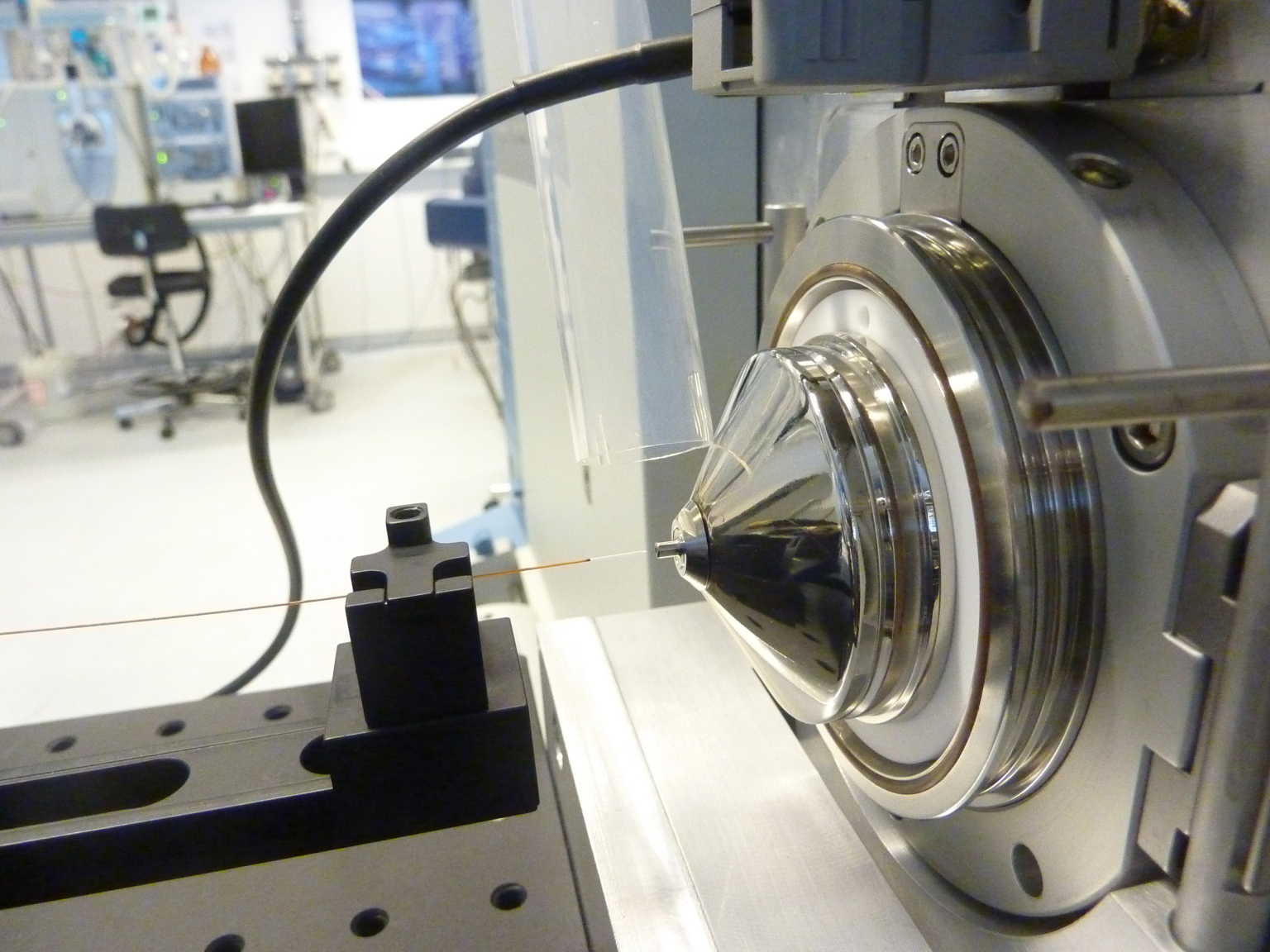
The PCF’s team supports proteomics-related projects, from experimental design, data acquisition, bioinformatics analysis to data interpretation. The Facility is equipped with State-of-the-Art Mass Spectrometry instrumentation operated by a highly experienced team. The team routinely scouts for advancements in proteomics and works to implement them to meet the users’ needs. Finally, the team is actively participating in consulting, teaching and training of students and researchers who are interested in the field of mass spectrometry-based proteomics.
Please contact us for project requests or general questions about Proteomics.
Working together